Introduction
It is a project I got based on research paper ‘An Unsupervised Learning Model for Deformable Medical Image Registration’. In this paper, authors introduced an algorithm for unsupervised n-D image registration. Image registration is an important task in image processing used in many ways. For this paper, image registration is working with multiple images to find out the changing between them. Their VoxelMorph CNN Architecture is based on UNet. To check the architecture, the following code can be used.
“import tensorflow as tf
tf.keras.utils.plot_model(vxm_model, show_shapes=True)”
They actually have two architectures. One is faster, the other one has higher accuracy. To understand better, the two input images are moving image(M) and fixed image(F). During the training, parameters for function g registers M through F. With spatial transform function, the output is moved image. Then loss function is used to check between moved image and fixed image.
Test with Heart Image
The training and testing code should be “train_miccai2018.py” and “test_miccai2018.py”. I don’t have a gpu on my laptop. Therefore, I just run the code on kaggle. After I run the code, I received the following image output. The first line is three slices from Fixed 3D Image(F). It is the second untreated brain image. Second line is Moved Image. It is the output after registration. They are nice and clean
To go deeper, I tried to test with some heart image. I only changed the input images and the parameters about images’ size. I used the same 3D model from the paper which was trainned from brain images. The size of input need to be multipled by 32. The brain image is [160,192,224]. The heart image I used is [64,32,32]. The output is showing in the following image. Yes, it is just part of the heart image. Also, the result doesn’t go very well. The registrated image disappeared. The grey levels are different.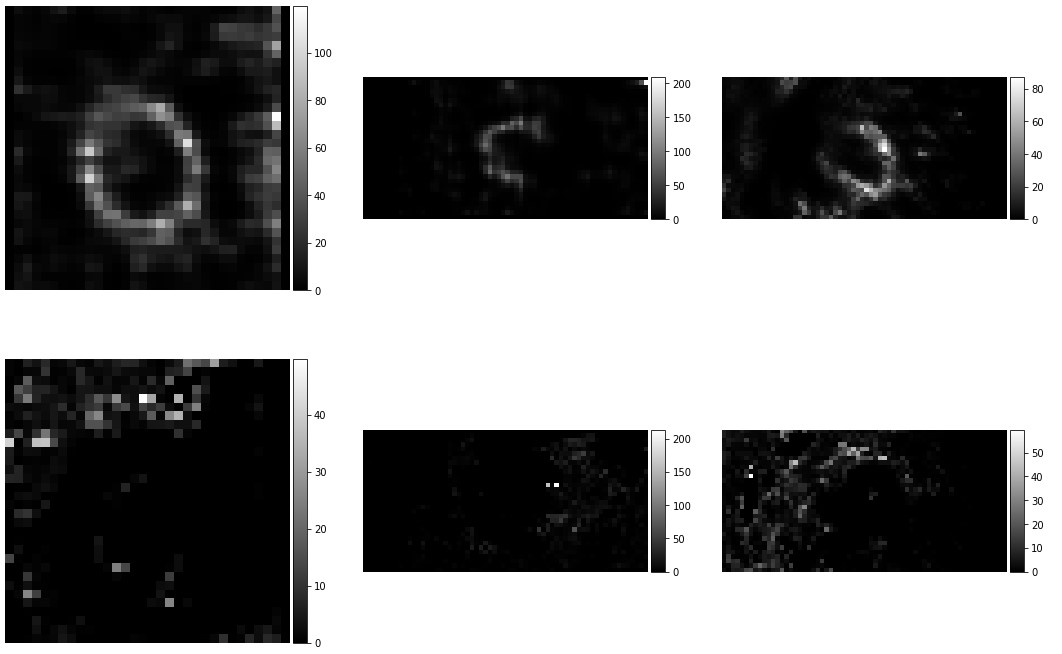
Next step, I am going to use 3D gaussian filter to denoise the heart images and check the plot function.
The plot function is “neuron.plot.slices(mid_slices_fixed + mid_slices_pred, cmaps=[‘gray’], do_colorbars=True, grid=[2,3]);” From how they import neuron, I find out the plot.py code under “voxelmorph-master/ext/neuron/neuron”. For gaussian filter, “scipy.ndimage” package is helpful.
To check the max element in an array: np.amax()
I resized the brain image to [64,32,32] to check whether the small size image cost the problem. Then I changed the orientation for the dimensionalities of images. At the end, I sloved the problem by change the grey level of heart images. The grey level for the brain image is under 1. Need to be careful for the input samples run in the test are the updated images.
Histogram for 3D Images
To make sure the heart images are scaled correctly, I need to find out 95% of largest value from their histograms. Then normalize to get new images and register the new images.
The heart images are really dark their histograms is similar with the following graph. PS. There are three or four voxels from the original images are above 800. I am not sure why, but I used gaussian filter. Then the maxmum for these arraies are below 255. img.astype(‘int’) can be used to change data type in array.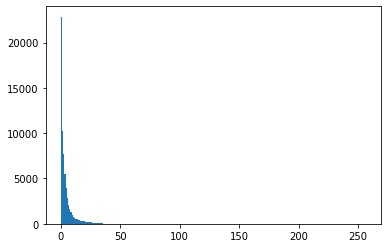
One mistake and updating
After contact with my professor, I noticed the output results are incorrect. The registrated images actually disappeared. The reason to cost this problem is I did not input the normalized images. After normalized, the gray level for images are all between 0 and 1. The following code is how I normalized all of these images.
max1 = np.amax(heart_image_1)
max2 = np.amax(heart_image_2)
min1 = np.amin(heart_image_1)
min2 = np.amin(heart_image_2)
print(max1,max2,min1,min2)
heart_image_1 = (heart_image_1-min1)/(max1-min1)
heart_image_2 = (heart_image_2-min2)/(max2-min2)
Instead of remove 95% of brightest pixels. I actually should only remove 1% of brightest pixels. The code I used is showing here:
greyLevel1 = heart_image_1.ravel()
greyLevel2 = heart_image_2.ravel()
newGrey1 = np.sort(-greyLevel1)
newGrey2 = np.sort(-greyLevel2)
[a,b,c] = heart_image_1.shape
for x in range(a):
for y in range(b):
for z in range(c):
if heart_image_1[x,y,z] > (-newGrey1[656]):
heart_image_1[x,y,z] = (-newGrey1[656])
for d in range(a):
for e in range(b):
for f in range(c):
if heart_image_2[d,e,f] > (-newGrey2[656]):
heart_image_2[d,e,f] = (-newGrey2[656])
Removing 1% brightest pixels VS. Gaussian filter
Next step, I compared two different ways to denoise heart images. One is removing 1% brightest pixels before normalized and registrated. The other one is using Gaussian filter to denoise before normalized and registrated. Package scipy.ndimage has 3D gaussian filter. With the package code is very simple:
from scipy.ndimage import gaussian_filter
heart_image_1 = gaussian_filter(heart_image_1,sigma=1)
heart_image_2 = gaussian_filter(heart_image_2,sigma=1)
The output shows gaussian filter has lower registrated error. But the images are fuzzier because it over denoised. Even though removing 1% brightest pixels has higher error data. It still is a better way for image processing.
Conclusion
It is a summer project I worked in 2020. For me, it is a chance understand image registration and try normalization, gaussian filter,etc. This project, itself, is not challengeable. The model was finished by others. It still is an enter level work for me to go through and study these topics.
References
https://openaccess.thecvf.com/content_cvpr_2018/papers/Balakrishnan_An_Unsupervised_Learning_CVPR_2018_paper.pdf
https://github.com/voxelmorph/voxelmorph
If you have any questions, please contact with tianluwu@gmail.com